Pablo Rodriguez-Mier

I am a research scientist at Saez-Rodriguez group (Institute for Computational Biomedicine, Heidelberg University) where I am currently working on biological network inference. I am interested in deciphering how complex biological systems behave under different conditions through mechanistic and computational models that integrate both prior biological knowledge and experimental data.
Prior to that, I was a postdoctoral researcher in Computational Systems Biology at INRAE Toxalim (Toulouse, France), where I was working on computational models to understand and predict the metabolic deregulation of cancer cells due to mutations in the TP53 gene.
news
| Mar 16, 2024 | Presenting CORNETO at EMBL Heidelberg |
|---|---|
| Mar 11, 2024 | I will be attending the EMBO-EMBL “AI and Biology” Symposium (Heidelberg) |
| Jan 18, 2024 | We won the Kaggle competition "Open Problems - Single-Cell Perturbations"! |
latest posts
| Mar 01, 2020 | Smart Invaders: Can You Beat Them? |
|---|---|
| Sep 05, 2017 | A tutorial on Differential Evolution with Python |
selected publications
- BioRxiv
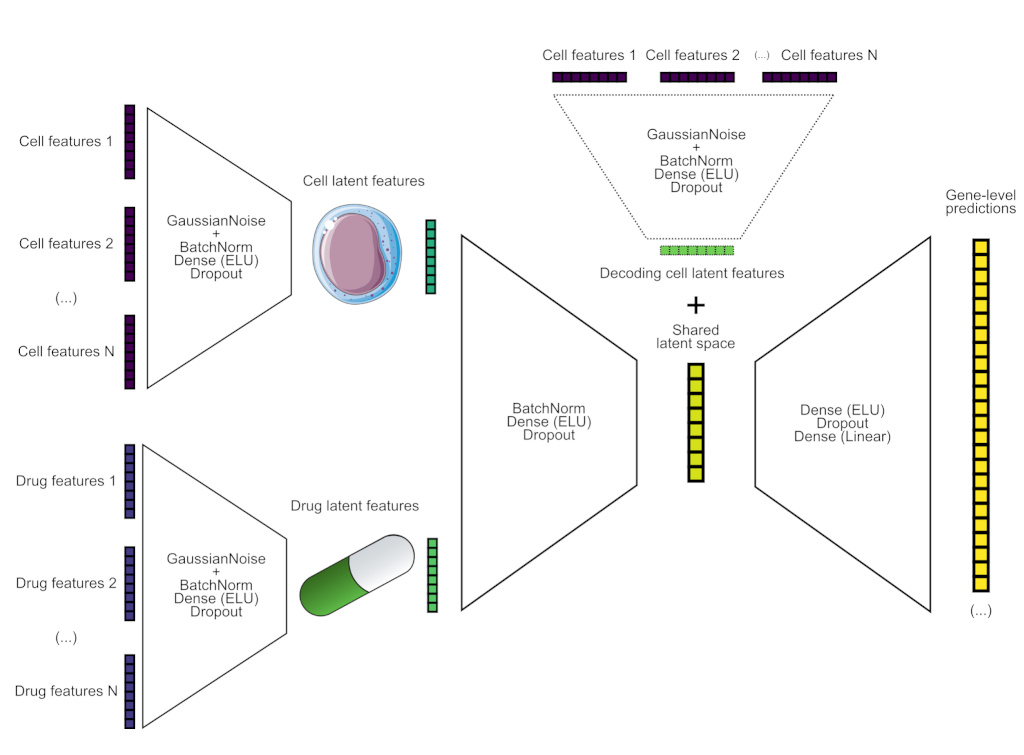 ScAPE: A lightweight multitask learning baseline method to predict transcriptomic responses to perturbationsbioRxiv, 2025
ScAPE: A lightweight multitask learning baseline method to predict transcriptomic responses to perturbationsbioRxiv, 2025 - Nat. Mach. Intell.
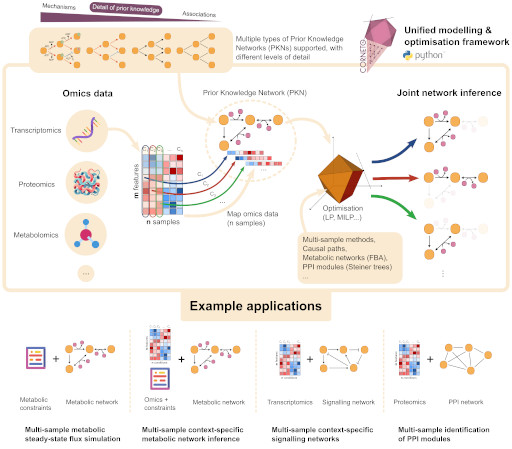 Unifying multi-sample network inference from prior knowledge and omics data with CORNETONature Machine Intelligence, 2025
Unifying multi-sample network inference from prior knowledge and omics data with CORNETONature Machine Intelligence, 2025 - Nat. Cell Biol.
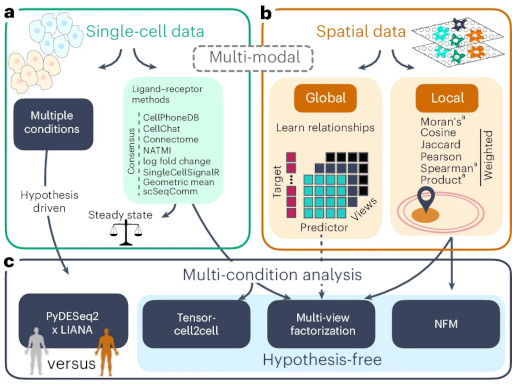 LIANA+ provides an all-in-one framework for cell–cell communication inferenceNature Cell Biology, 2024
LIANA+ provides an all-in-one framework for cell–cell communication inferenceNature Cell Biology, 2024 - Bioinformatics
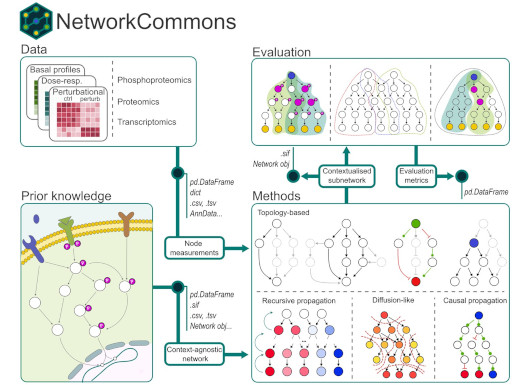 NetworkCommons: bridging data, knowledge, and methods to build and evaluate context-specific biological networksBioinformatics, 2025
NetworkCommons: bridging data, knowledge, and methods to build and evaluate context-specific biological networksBioinformatics, 2025 - PLoS Comput. Biol.
 DEXOM: Diversity-based enumeration of optimal context-specific metabolic networksPLoS Computational Biology, 2021
DEXOM: Diversity-based enumeration of optimal context-specific metabolic networksPLoS Computational Biology, 2021 - Comput. Biomed.
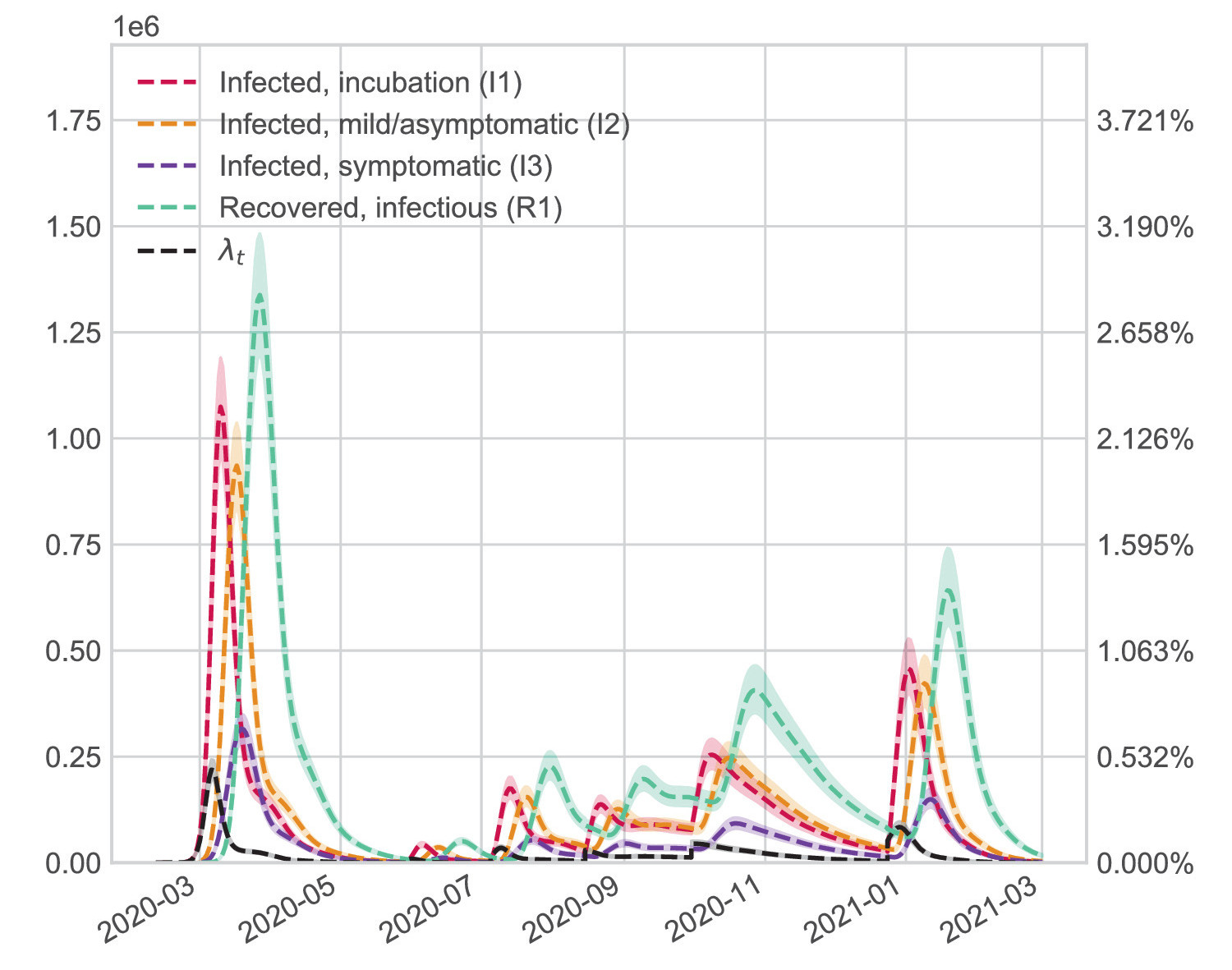 COVID-19: Estimation of the transmission dynamics in Spain using a stochastic simulator and black-box optimization techniquesComputer Methods and Programs in Biomedicine, 2021
COVID-19: Estimation of the transmission dynamics in Spain using a stochastic simulator and black-box optimization techniquesComputer Methods and Programs in Biomedicine, 2021
